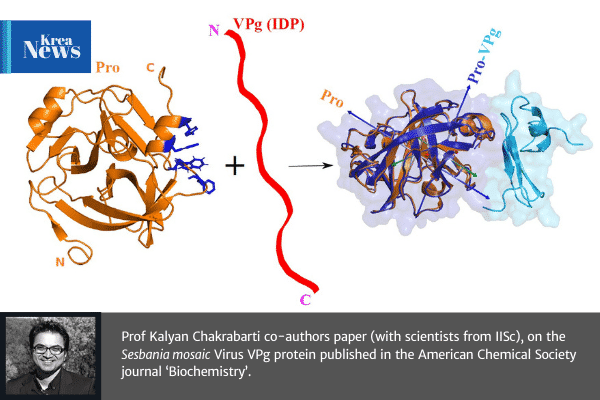
Prof Kalyan Chakrabarti – Assistant Professor of Biological Science & Chemistry, Krea University – co-authors with scientists at IISc for a paper on “Aromatic Interactions Drive the Coupled Folding and Binding of the Intrinsically Disordered Sesbania mosaic Virus VPg Protein” published in the American Chemical Society journal ‘Biochemistry’.
About the paper:
The cells are the tiny units of life. Protein molecules are the engines that carry out all the necessary work within the cell. But, how are the engines assembled within the cell? A team of scientists, including Prof Kalyan Chakrabarti, have answered this question in an article published in the American Chemical Society journal ‘Biochemistry’. The work, partially funded by the Krea Intramural Fellowship awarded in 2019, discusses a specific problem of viral infection in plants which has the potential to provide a blueprint for preventing viral infections in animals too.
Abstract:
The plant Sesbania mosaic virus [a (+)-ssRNA sobemovirus] VPg protein is intrinsically disordered in solution. For the virus life cycle, the VPg protein is essential for replication and for polyprotein processing that is carried out by a virus-encoded protease. The nuclear magnetic resonance (NMR)-derived tertiary structure of the protease-bound VPg shows it to have a novel tertiary structure with an α-β-β-β topology. The quaternary structure of the high-affinity protease–VPg complex (≈27 kDa) has been determined using HADDOCK protocols with NMR (residual dipolar coupling, dihedral angle, and nuclear Overhauser enhancement) restraints and mutagenesis data as inputs. The geometry of the complex is in excellent agreement with long-range orientational restraints such as residual dipolar couplings and ring-current shifts. A “vein” of aromatic residues on the protease surface is pivotal for the folding of VPg via intermolecular edge-to-face π···π stacking between Trp271 and Trp368 of the protease and VPg, respectively, and for the CH···π interactions between Leu361 of VPg and Trp271 of the protease. The structure of the protease–VPg complex provides a molecular framework for predicting sites of important posttranslational modifications such as RNA linkage and phosphorylation and a better understanding of the coupled folding upon binding of intrinsically disordered proteins. The structural data presented here augment the limited structural data available on viral proteins, given their propensity for structural disorder.
Reference: Karuna Dixit, N. Megha Karanth, Smita Nair, Khushboo Kumari, Kalyan S. Chakrabarti, Handanahal S. Savithri, and Siddhartha P. Sarma | Biochemistry 2020 59 (49), 4663-4680 | DOI: 10.1021/acs.biochem.0c00721
Read the complete paper, here.